The intricate processes that govern embryonic body patterning have been elucidated by researchers at RIKEN, a leading Japanese research institute. Their recent studies reveal how specific genes orchestrate the formation of somites, the structures that eventually develop into the backbone, ribs, and muscles. This groundbreaking work sheds light on the molecular mechanisms behind these developmental processes.
The formation of somites occurs through a cyclical activation of a network of genes, an event driven by what is known as the segmentation clock. While many genes involved in this process have been identified, the precise molecular steps that result in synchronized gene activity remain unclear. According to Ryoichiro Kageyama of the RIKEN Center for Biosystems Dynamics Research, the dynamics of these oscillations propagate like waves across embryonic tissue, making their study challenging.
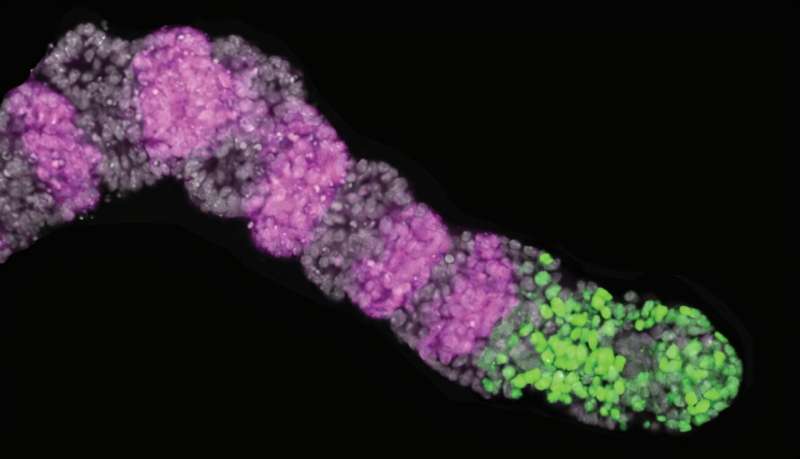
To address this complexity, Kageyama and his team have leveraged advanced technologies to track and manipulate somite formation in real time. They engineered mouse stem cells to develop into induced presomitic mesoderm (iPSM), a precursor tissue that closely resembles the embryonic environment where somites form. Their findings are documented in two significant articles published in the journals Development and Genes & Development.
Kageyama highlights that these iPSM models maintain essential characteristics of the in vivo segmentation clock. This technological advancement enabled the researchers to analyze the role of a key oscillator gene, Hes7, in somite formation. Specifically, Hes7 influences the expression of downstream genes, including Cdh2, which encodes an adhesion protein critical for cellular bonding.
The research indicates that Hes7’s regulation of Cdh2 plays a vital role in the signaling pathways that direct the timing of cell maturation during somite segmentation. By manipulating gene expression within the iPSM constructs and employing live-cell microscopy, the team demonstrated that alterations in Cdh2 activity, resulting from disrupted Hes7 expression, led to irregularities in cell adhesion. This, in turn, compromised the structural integrity of developing somites.
Kageyama emphasizes the significance of these findings, stating, “This discovery provides another molecular bridge between oscillatory gene expression and signaling pathways, revealing how temporal information is translated into spatial patterns during embryogenesis.” This work represents a crucial step toward the researchers’ goal of creating a comprehensive model that reconstructs the spatial and temporal dynamics of somite formation.
The studies published by Kageyama’s team reflect a significant advancement in understanding the biological mechanisms that shape vertebrate development, promising to inform future research in developmental biology and regenerative medicine. The findings may also have broader implications for understanding congenital malformations and other developmental disorders.
For more detailed insights, the full research articles can be accessed in the journals Development (DOI: 10.1242/dev.204743) and Genes & Development (DOI: 10.1101/gad.352538.124).