UPDATE: A groundbreaking study published on 5 July 2025 by a team from KU Leuven has unveiled a revolutionary deep learning model that dramatically enhances the analysis of fruit tissue microstructure. This innovative approach outperforms traditional 2D methods, achieving remarkable accuracy in the 3D imaging of apples and pears.
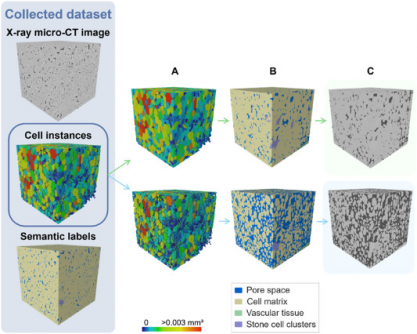
The new model addresses the limitations of conventional microscopy techniques, which demand extensive sample preparation and provide limited fields of view. X-ray micro-CT now allows for non-destructive 3D imaging, but quantifying tissue morphology has remained a challenge due to overlapping features and low image contrast. The study introduces an automated framework capable of accurately segmenting complex fruit tissues, which could transform research in plant physiology and storage behavior.
Using a 3D panoptic segmentation framework based on the Cellpose model and a 3D Residual U-Net, the research team successfully labeled and quantified the microstructure of fruit tissues. The model executed both instance and semantic segmentation, effectively distinguishing between parenchyma cells, vascular tissues, and stone cell clusters. The results were benchmarked against a 2D segmentation model and a traditional marker-based watershed algorithm.
Evaluation metrics revealed that the 3D model achieved an Aggregated Jaccard Index (AJI) of 0.889 for apples and 0.773 for pears, surpassing the 2D model’s performance of 0.861 and 0.732 respectively. The model showcased nearly perfect segmentation of pore spaces and cell matrices, while identifying vascular structures in apples with a Dice Similarity Coefficient (DSC) of 0.506 and 0.789 in pears.
Visual validation confirmed the model’s accuracy, successfully detecting vascular bundles in ‘Kizuri’ and ‘Braeburn’ apples and providing realistic segmentation of stone cell clusters in ‘Celina’ and ‘Fred’ pears, with DSC scores reaching up to 0.90. However, challenges remain; additional data augmentation did not improve performance, likely due to dataset imbalance.
This pioneering model represents a significant advancement in the field, offering plant scientists a powerful, non-destructive tool to study how microscopic structures impact nutrient transport, texture, and storability of fruit. It simplifies the “human-in-the-loop” analysis process, reducing the need for manual labor while enhancing accuracy in tissue characterization.
Moreover, the technology presents a scalable framework for investigating tissue development, ripening, and stress responses across various crops. Its compatibility with standard X-ray micro-CT instruments makes it an accessible solution for integrating artificial intelligence into plant anatomy and food science research.
As this technology continues to evolve, the implications for agriculture and food science are profound, potentially leading to better crop management strategies and improved food quality. Researchers and industry professionals are encouraged to explore this innovative approach to deepen their understanding of plant tissue microstructures and enhance agricultural practices.
For further details, the study is published in Plant Phenomics (DOI: 10.1016/j.plaphe.2025.100087).