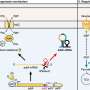
Recent research has revealed that a small RNA derived from the 3′ untranslated region (3′UTR) plays a critical role in linking acid resistance with metabolic reprogramming in the bacterium Salmonella while it resides within macrophages. This finding enhances the understanding of how Salmonella, a facultative intracellular pathogen, survives in hostile environments, such as the acidic conditions found in the gastrointestinal tract and macrophage phagosomes.
Acid resistance is vital for enterobacteria, enabling them to endure the low pH levels encountered during infection. The study highlights the function of the enzyme arginine decarboxylase AdiA, which is crucial for Salmonella to thrive in acidic environments. AdiA operates by catalyzing a reaction that consumes protons (H+), thereby contributing to the bacterium’s survival strategies.
Understanding the Mechanisms of Survival
The research underscores the importance of metabolic adaptations in Salmonella as it navigates acidic environments. The small RNA identified in the study appears to facilitate communication between acid resistance mechanisms and metabolic pathways. This connection enables Salmonella to efficiently utilize resources within macrophages, ensuring its survival and replication.
During infection, Salmonella not only confronts the acidic conditions but also must evade the immune response. The ability to alter its metabolism in response to environmental changes is a key factor in its pathogenicity. The study’s findings suggest that the interplay between acid resistance and metabolic reprogramming may offer new avenues for therapeutic intervention against Salmonella infections.
Researchers employed advanced molecular techniques to uncover how this small RNA influences the expression and activity of AdiA. This insight provides a deeper understanding of the mechanisms by which Salmonella adapts to the challenging conditions within host cells.
Implications for Future Research
The implications of these findings extend beyond basic microbiology. Understanding the metabolic adaptations of Salmonella could inform the development of novel strategies to combat infections caused by this pathogen. By targeting the specific pathways involved in acid resistance and metabolic reprogramming, researchers may be able to devise new treatments that disrupt the bacterium’s survival mechanisms.
Further studies will be necessary to explore the potential of these findings in clinical applications. As the research community continues to investigate the dynamics of Salmonella within host cells, the hope is to gain insights that could lead to effective preventive measures and therapies against enterobacterial infections.
In summary, the connection between acid resistance and metabolic reprogramming in Salmonella highlights the complexity of microbial survival strategies in hostile environments. The ongoing research efforts will be crucial in understanding how to counteract such pathogens effectively.